At present, the breakthrough development of DNA sequencing technology is driving unprecedented systematic changes in the agricultural field. Since the sequencing of the first plant genome, the iterative upgrading of molecular biology technology has continuously expanded the cognitive boundary of agricultural science: from the fine positioning of key agronomic traits genes to the successful practice of targeted genome editing technology based on CRISPR-Cas9 system in crop improvement, the modern agricultural system is accelerating the transformation from the extensive mode dominated by traditional experience to the precise and intelligent paradigm relying on genomic big data.
This paper expounds the application of DNA sequencing technology in agricultural practice, and shows its role in promoting the transformation of agriculture to precision intelligence paradigm.
DNA Sequencing in Modern Crop Breeding
The completion of the rice genome sketch in 2000 marked the entry of crop breeding into the genome era. As the staple food of half of the world's population, the genome sequence analysis of rice with 430 million base pairs provides a molecular basis for identifying key traits such as yield and stress resistance. Subsequently, the whole genome sequencing of corn, wheat, soybean and other important crops was completed one after another, which enabled breeders to understand the genetic mechanism of trait formation at the DNA level.
The progress of genome sequencing technology has promoted the efficiency revolution of crop breeding. Early map sequencing relied on BAC cloning and Sanger sequencing, which took several years and was expensive. However, the second generation sequencing (NGS) technology reduced the cost of rice genome resequencing to less than 1,000 dollars and shortened the time to several weeks. Third-generation single-molecule long reading and long sequencing (such as PacBio SMRT) has solved the assembly problem of complex genomes (such as hexaploid wheat), completed the fine map of 21 pairs of chromosomes in wheat, and made it possible to clone complex trait genes such as powdery mildew resistance gene Pm21.

Overview of three high-throughput next generation sequencing systems (Huq et al., 2016)
In traditional breeding, disease resistance screening depends on field inoculation identification, which is time-consuming and laborious and easily affected by the environment. Marker-assisted selection (MAS) technology driven by DNA sequencing completely revolutionized this situation. Based on the in-depth analysis of crop genome, this technology accurately locates molecular markers closely linked with target traits by means of genome-wide association study (GWAS) and linkage analysis.
In the field of drought-resistant breeding, DNA sequencing is becoming the key to unlock the survival code of plants in adversity. Through GWAS and transcriptome sequencing technology, researchers have successfully mapped the molecular regulatory network of plants in response to drought stress. The core advantage of MAS technology lies in the ability of "phenotypic prediction". Compared with traditional breeding, which needs to wait for the whole growth cycle to observe the performance of traits, MAS technology can infer the potential phenotype of plants at seedling stage or even seed stage by detecting DNA markers closely linked to target traits. This feature is particularly significant in improving quantitative traits (such as yield and quality) controlled by multiple genes, shortening the traditional breeding cycle from 8-10 years to 3-5 years, and avoiding the interference of environmental factors on phenotypic identification, thus improving the breeding efficiency by 3-5 times.

Schematic workflow for marker assay development (Mori et al., 2023)
DNA Sequencing Empowering Transgenic and Gene-Edited Crop Innovation
As an important branch of modern biotechnology, transgenic technology improves crop traits by introducing foreign genes, and DNA sequencing technology provides an indispensable molecular biological basis for gene cloning and expression regulation mechanism analysis. Taking the research and development of insect-resistant cotton as an example, researchers successfully isolated functional gene fragments encoding Cry1Ac protein by sequencing the genes of Bacillus thuringiensis (Bt). With the help of Agrobacterium-mediated genetic transformation system, the gene was stably integrated into cotton genome, and the field test data showed that its mortality rate to cotton bollworm reached over 95%.
As a revolutionary breakthrough in the field of modern molecular biology, gene editing technology realizes character improvement through targeted modification of crop genome. Among them, CRISPR-Cas9 system has become the core tool of plant gene editing because of its high specificity, simple operation and multi-species applicability.
Taking rice (Oryza sativa) as the research object, editing the promoter region of OsSPL14 gene can up-regulate the expression level of the gene, and by adjusting the tillering development mode of the plant, the number of grains per panicle per unit area is significantly increased, and finally the yield is increased by about 15%. Directional transformation of OsEPSPS gene endows rice with glyphosate resistance, which provides a new path for intensive farmland management.

Overview of NGS applications in crop genetics and breeding (Varshney et al., 2009)
DNA Sequencing in Livestock Breeding Optimization
In the process of modern agricultural development, livestock and poultry optimization achieved a major breakthrough with the help of genome selection technology. Based on DNA sequencing, this technology accurately analyzes the genetic information of livestock and poultry, drives the breeding of varieties in the direction of high yield and low consumption, completely changes the traditional breeding mode, and provides a powerful driving force for ensuring the supply of livestock and poultry products and improving industrial benefits.
Genome Selection and Meat Quality Improvement in Pigs
DNA sequencing technology has completely changed the pig breeding mode. In traditional breeding, important economic traits such as lean meat percentage and feed conversion rate need to be determined after slaughter, while genome selection (GS) predicts individual breeding value through genome-wide SNP markers, which greatly improves the breeding efficiency. The genome sequencing of domestic pigs completed in 2009 showed that its 2.7 Gb genome contained 22,000 protein coding genes, which provided abundant molecular markers for GS.
In the aspect of meat quality improvement, DNA sequencing technology has brought a revolutionary breakthrough for pig breeding. Through GWAS, scientists have successfully located several key gene loci affecting pork quality. Among them, the c.1843CT mutation of RYR1 gene (commonly known as halothane gene) is like a bomb. Pigs carrying this mutation will trigger an abnormal calcium release mechanism under stress, which will lead to continuous muscle contraction and energy depletion, and eventually form PSE meat (pale meat, soft texture and serious water seepage).
According to statistics, under the traditional breeding mode, the incidence of PSE meat in pigs carrying the mutant gene is as high as 30%-40%, but after the carriers are accurately identified and eliminated by DNA detection technology, this proportion can drop to below 5%. The c.307GA mutation of FUT1, another key gene, showed unique disease resistance advantages. It was found that the mutation could change the receptor structure on the surface of intestinal cells, which made E.coli F18 unable to adhere effectively, thus significantly reducing the risk of diarrhea in weaned piglets.
 Accuracy of GEBV by observed accuracy of EBV (Badke et al., 2014)
Accuracy of GEBV by observed accuracy of EBV (Badke et al., 2014)
Selection of Bovine Genome and Improvement of Milk Meat Quality
Genome selection has become the mainstream technology in dairy cattle breeding. As the highest milk yield dairy cow breed in the world, the whole genome sequencing of Holstein dairy cows revealed that the key economic traits such as milk yield, milk fat rate and milk protein rate were co-regulated by hundreds of minor genes, which were distributed on 22 pairs of autosomes and sex chromosomes. It is difficult for traditional breeding methods to capture the superposition effect of minor genes, but the emergence of high-density SNP chip technology has completely changed this situation.
By constructing a reference population containing tens of thousands of cows, the genome breeding value (GEBV) of breeding bulls was calculated by using GWAS and genome optimal linear unbiased prediction (GBLUP) algorithms. The data show that the genetic progress of milk yield is increased from 100 kg to 150 kg per year in traditional breeding, which is equivalent to a 50% increase in genetic improvement efficiency every year. At the same time, the generation interval was greatly shortened from 6 years to 3 years, which significantly accelerated the process of genetic improvement.
In the field of beef cattle breeding, DNA sequencing technology is reshaping the traditional breeding model and becoming the core driving force for cultivating high-quality beef cattle breeds. Using GWAS, the researchers successfully located the key gene CAPN1 which affects beef tenderness. The calcium-activated neutral protease encoded by this gene plays an important role in the process of meat ripening after slaughter. Through in-depth study of CAPN1 gene, scientists have developed functional markers with high accuracy, which can quickly identify breeding cattle with excellent tenderness traits. After the application of this technology, the qualification rate of meat tenderness in Australian beef cattle industry increased by 25%, which significantly enhanced the competitiveness of products in the international market.

Timeline of an aggressive artifi cial insemination breeding program based on the use of genomic bulls as sires of sons (Schefers et al., 2012)
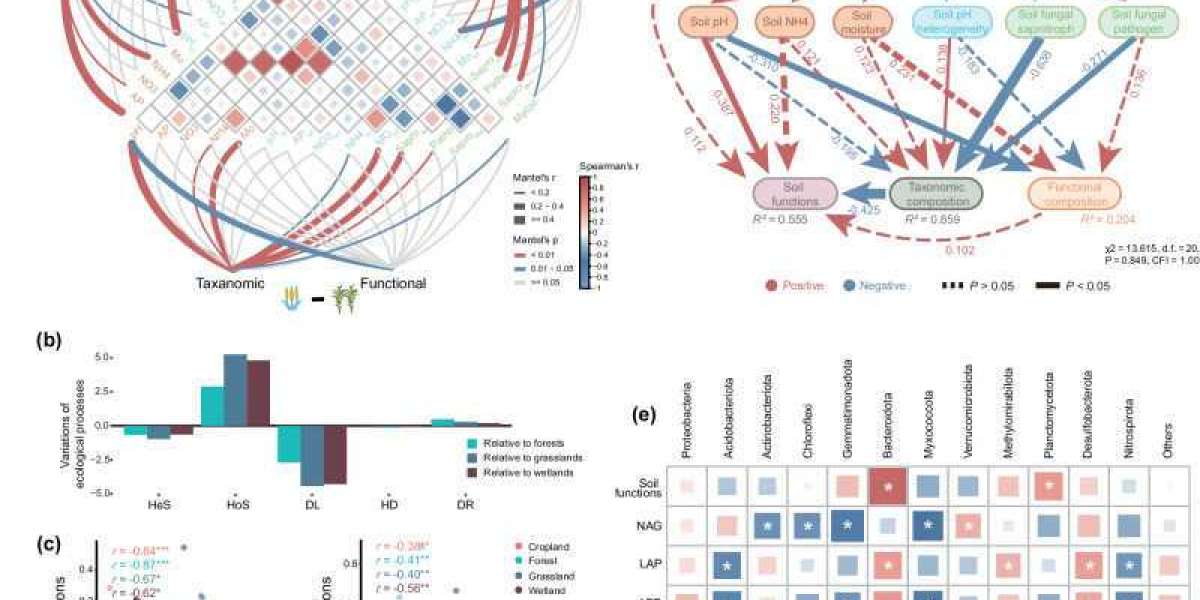
DNA Sequencing for Soil Microbiome and Crop Health
There is a complex interaction between soil microbial community structure and crop health, and the analysis of its internal mechanism is the key to realize the sustainable development of agriculture. In recent years, metagenomics technology based on high-throughput sequencing has become an important tool to analyze soil-crop microbiota with its high resolution and wide coverage.
Rhizosphere Microbial Sequencing and Ecological Agriculture
As the "second genome" of crop growth, soil microbiota plays an irreplaceable role in plant health regulation. DNA sequencing technology reveals the complex interaction mechanism between soil microbial community structure and crop health by analyzing it. High-throughput sequencing technology with 16S rRNA gene sequencing as the core can systematically analyze rhizosphere microbial communities.
It is found that healthy soil is usually rich in beneficial bacteria such as actinomycetes and Bacillus, which can help crops resist the invasion of pathogenic bacteria by producing antibiotics, plant hormones or inducing systemic resistance. In the continuous cropping obstacle soil, Fusarium, Rhizoctonia solani and other pathogens will gradually occupy a dominant position, causing soil-borne diseases.
Functional metagenomics technology, with its unique molecular biology perspective, opens a new window for analyzing the functional potential of soil microbiota. Using advanced DNA extraction technology, researchers isolated the total DNA of microorganisms from black soil, and successfully identified several key functional gene clusters through Qualcomm quantitative metagenome sequencing. In these gene resource banks:
- Organophosphorus degradation genes can transform organophosphorus in soil that is difficult to be absorbed by crops into available phosphorus forms, which significantly improves the utilization rate of phosphate fertilizer.
- Nitrogen-fixing genes drive microorganisms to convert nitrogen in the air into ammonia nitrogen available to plants, reducing dependence on chemical nitrogen fertilizer.
- The metabolic pathway encoded by plant hormone synthesis genes can regulate the balance of crop growth hormone under adverse conditions and enhance its resistance to drought and cold.

Environmental drivers of microbial composition and their relationship to soil functions at continental scale (Peng et al., 2024)
egulation of Microbe and Sustainable Agriculture
Microorganism regulation technology under the guidance of DNA sequencing is becoming a new way of sustainable agriculture. 16S rRNA gene analysis based on high-throughput sequencing platform can analyze soil microbial community structure with single base accuracy. In the rice-duck cooperative ecosystem, sequencing data revealed that ducks reshaped the niche distribution of microorganisms in rice fields by eating weeds and stirring water bodies.
Sequencing data show that this symbiotic model can increase the relative abundance of AOA by 40%-55% and the number of denitrifying bacteria by 2-3 orders of magnitude. The dynamic change of microbial community effectively inhibited the activity of methanogenic bacteria, reduced the methane emission intensity of rice fields by 25-30%, and improved the nitrogen utilization rate of soil by 15% by strengthening nitrogen cycle.
Microbial sequencing provides a new strategy for crop disease prevention and control. In traditional agricultural control, the high incidence of Fusarium wilt caused by cucumber continuous cropping has always been a difficult problem for farmers. Pathogens accumulate in the soil year by year, and chemical pesticide control is not only costly, but also easy to cause environmental pollution and pesticide residues.
With the development of microbial sequencing technology, the research team deeply analyzed the microbial community structure of cucumber continuous cropping soil, and screened out the core flora with antagonistic effect to Fusarium wilt through high-throughput sequencing. Among them, Bacillus can secrete lipopeptide antibiotics and inhibit the mycelium growth of pathogenic bacteria; Trichoderma directly decomposes pathogenic bacteria cells through heavy parasitism; Actinomycetes can produce a variety of antibacterial active substances, forming multiple lines of defense.

Plant-microbial interactions and sustainable agriculture applications (Sudheer et al., 2020)
Conclusion
The analysis of crop genome based on Qualcomm DNA sequencing technology, the iteration of molecular breeding technology system and the transformation of agricultural production paradigm are reshaping the interaction between human and cultivated land. Breakthrough progress has been made in the fields of crop character improvement mediated by gene editing technology (such as CRISPR/Cas system), directional regulation of soil microbiome, and genome selection and breeding of livestock and poultry, which not only highlights the core position of biotechnology in modern agriculture, but also reflects the strategic thinking of human beings to ensure food security by using life science theory.
The core value of this technological innovation lies in breaking through the limitations of traditional empirical agriculture. By integrating genomic data, molecular design and breeding and precision agriculture technology, the synergy between crop yield improvement, quality optimization and sustainable development of ecological environment can be realized, and biological solutions can be provided to global challenges such as climate change and population growth.